| 质粒名称: | Circular-ic-EMCV-GFP |
|---|---|
| 出品公司: | Addgene |
| 目录编号: | 226259 |
| 存储实验室: | Prashant Mali |
|
载体骨架基本信息 |
|
| 载体名称: | 未知 |
| 载体出品公司: | Clontech |
| 空载体大小: | 5485 bp |
| 完整质粒大小: | 5485 bp |
| 载体类型: | In Vitro Transcription |
| 载体抗性: | 氨苄霉素 |
| 生长温度: | 37 ℃ |
| 宿主菌: | DH5a |
| 拷贝数: | 高拷贝 |
|
插入基因信息 |
|
| 插入基因名称: | EGFP |
| 物种来源: | Synthetic |
| 克隆方法: | Gibson Cloning |
|
文献引用方法 |
|
| 材料和方法部分: | Circular-ic-EMCV-GFP was a gift from Prashant Mali (Addgene plasmid # 226259 ; http://n2t.net/addgene:226259 ; RRID:Addgene_226259) |
| 参考文献部分: | Robust genome and cell engineering via in vitro and in situ circularized RNAs. Tong M, Palmer N, Dailamy A, Kumar A, Khaliq H, Han S, Finburgh E, Wing M, Hong C, Xiang Y, Miyasaki K, Portell A, Rainaldi J, Suhardjo A, Nourreddine S, Chew WL, Kwon EJ, Mali P. Nat Biomed Eng. 2024 Aug 26. doi: 10.1038/s41551-024-01245-z. 10.1038/s41551-024-01245-z PubMed 39187662 |
买家导航

Circular-ic-EMCV-GFP载体质粒基本信息
Circular-ic-EMCV-GFP质粒图谱载体图谱和Circular-ic-EMCV-GFP载体序列质粒序列多克隆位点信息
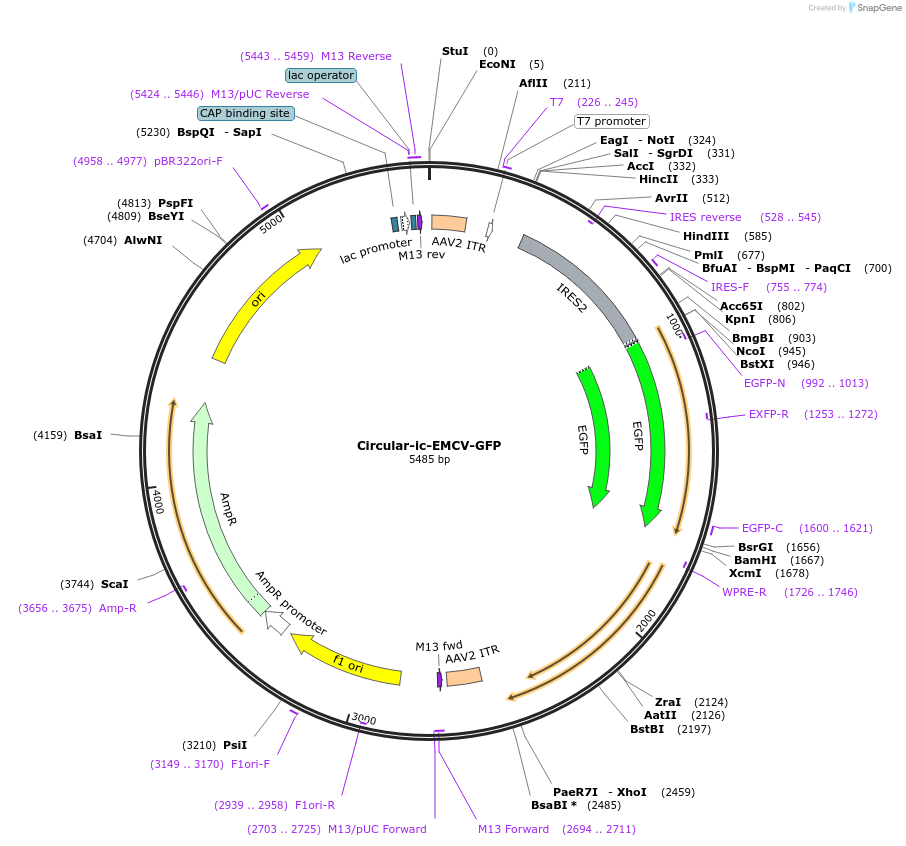
Circular-ic-EMCV-GFP质粒载体简介
Circular-ic-EMCV-GFP质粒序列载体序列
LOCUS Circular-ic-EMCV-GFP 5485 bp DNA circular SYN 14-APR-2025
DEFINITION Production of in situ circularizable RNA via IVT.
ACCESSION .
VERSION .
KEYWORDS .
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 5485)
AUTHORS Tong M, Palmer N, Dailamy A, Kumar A, Khaliq H, Han S, Finburgh E,
Wing M, Hong C, Xiang Y, Miyasaki K, Portell A, Rainaldi J, Suhardjo
A, Nourreddine S, Chew WL, Kwon EJ, Mali P
TITLE Robust genome and cell engineering via in vitro and in situ
circularized RNAs.
JOURNAL Nat Biomed Eng. 2024 Aug 26. doi: 10.1038/s41551-024-01245-z.
PUBMED 39187662
REFERENCE 2 (bases 1 to 5485)
AUTHORS .
TITLE Direct Submission
JOURNAL Exported Apr 14, 2025 from SnapGene Server 8.0.1
https://www.biofeng.com
COMMENT SGRef: number: 1; type: "Journal Article"; journalName: "Nat Biomed
Eng. 2024 Aug 26. doi: 10.1038/s41551-024-01245-z."
COMMENT Sequence Label: Circular-ic-EMCV-GFP
FEATURES Location/Qualifiers
source 1..5485
/mol_type="other DNA"
/organism="synthetic DNA construct"
repeat_region 12..141
/label=AAV2 ITR
primer_bind 226..245
/label=T7
/note="T7 promoter, forward primer"
promoter 226..244
/label=T7 promoter
/note="promoter for bacteriophage T7 RNA polymerase"
misc_feature join(361..934,935..937,938..946)
/label=IRES2
/note="internal ribosome entry site (IRES) of the
encephalomyocarditis virus (EMCV)"
primer_bind complement(528..545)
/label=IRES reverse
/note="IRES internal ribosome entry site, reverse primer.
Also called pCDH-rev"
primer_bind 755..774
/label=IRES-F
/note="IRES internal ribosome entry site, forward primer"
CDS join(947..949,950..952,953..1666)
/codon_start=1
/product="the original enhanced GFP (Yang et al., 1996)"
/label=EGFP
/note="mammalian codon-optimized"
/translation="MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTL
KFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDD
GNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIK
VNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLL
EFVTAAGITLGMDELYK"
CDS join(947..949,950..952,953..1663)
/codon_start=1
/product="enhanced GFP"
/label=EGFP
/translation="MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTL
KFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDD
GNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIK
VNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLL
EFVTAAGITLGMDELYK"
primer_bind complement(992..1013)
/label=EGFP-N
/note="EGFP, reverse primer"
primer_bind complement(1253..1272)
/label=EXFP-R
/note="For distinguishing EGFP variants, reverse primer"
primer_bind 1600..1621
/label=EGFP-C
/note="EGFP, forward primer"
primer_bind complement(1726..1746)
/label=WPRE-R
/note="WPRE, reverse primer"
repeat_region 2546..2675
/label=AAV2 ITR
primer_bind complement(2694..2711)
/label=M13 Forward
/note="In lacZ gene. Also called M13-F20 or M13 (-21)
Forward"
primer_bind complement(2694..2710)
/label=M13 fwd
/note="common sequencing primer, one of multiple similar
variants"
primer_bind complement(2703..2725)
/label=M13/pUC Forward
/note="In lacZ gene"
rep_origin 2852..3307
/direction=RIGHT
/label=f1 ori
/note="f1 bacteriophage origin of replication; arrow
indicates direction of (+) strand synthesis"
primer_bind complement(2939..2958)
/label=F1ori-R
/note="F1 origin, reverse primer"
primer_bind 3149..3170
/label=F1ori-F
/note="F1 origin, forward primer"
promoter 3333..3437
/gene="bla"
/label=AmpR promoter
CDS join(3438..3506,3507..4298)
/codon_start=1
/gene="bla"
/product="beta-lactamase"
/label=AmpR
/note="confers resistance to ampicillin, carbenicillin, and
related antibiotics"
/translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGYI
ELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRIDAGQEQLGRRIHYSQNDLVEYS
PVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRLDRW
EPELNEAIPNDERDTTMPVAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPLLRSA
LPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIAEIGAS
LIKHW"
primer_bind complement(3656..3675)
/label=Amp-R
/note="Ampicillin resistance gene, reverse primer"
rep_origin 4469..5057
/direction=RIGHT
/label=ori
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
primer_bind 4958..4977
/label=pBR322ori-F
/note="pBR322 origin, forward primer"
protein_bind 5345..5366
/label=CAP binding site
/bound_moiety="E. coli catabolite activator protein"
/note="CAP binding activates transcription in the presence
of cAMP."
promoter join(5381..5386,5387..5404,5405..5411)
/label=lac promoter
/note="promoter for the E. coli lac operon"
protein_bind 5419..5435
/label=lac operator
/bound_moiety="lac repressor encoded by lacI"
/note="The lac repressor binds to the lac operator to
inhibit transcription in E. coli. This inhibition can be
relieved by adding lactose or
isopropyl-beta-D-thiogalactopyranoside (IPTG)."
primer_bind 5424..5446
/label=M13/pUC Reverse
/note="In lacZ gene"
primer_bind 5443..5459
/label=M13 rev
/note="common sequencing primer, one of multiple similar
variants"
primer_bind 5443..5459
/label=M13 Reverse
/note="In lacZ gene. Also called M13-rev"
ORIGIN
1 ccttaattag gctgcgcgct cgctcgctca ctgaggccgc ccgggcaaag cccgggcgtc
61 gggcgacctt tggtcgcccg gcctcagtga gcgagcgagc gcgcagagag ggagtggcca
121 actccatcac taggggttcc ttgtagttaa tgattaaccc gccatgctac ttatctacgt
181 agccatgctc taggaagatc ggaattcgcc cttaagctag gatgctaata cgactcacta
241 tagggccatc agtcgccggt cccaagcccg gataaaatgg gagggggcgg gaaaccgcct
301 aaccatgccg actgatggca gagcggccgc gtcgacgggc ccgcggaatt ccgccccccc
361 cccctctccc tccccccccc ctaacgttac tggccgaagc cgcttggaat aaggccggtg
421 tgcgtttgtc tatatgttat tttccaccat attgccgtct tttggcaatg tgagggcccg
481 gaaacctggc cctgtcttct tgacgagcat tcctaggggt ctttcccctc tcgccaaagg
541 aatgcaaggt ctgttgaatg tcgtgaagga agcagttcct ctggaagctt cttgaagaca
601 aacaacgtct gtagcgaccc tttgcaggca gcggaacccc ccacctggcg acaggtgcct
661 ctgcggccaa aagccacgtg tataagatac acctgcaaag gcggcacaac cccagtgcca
721 cgttgtgagt tggatagttg tggaaagagt caaatggctc tcctcaagcg tattcaacaa
781 ggggctgaag gatgcccaga aggtacccca ttgtatggga tctgatctgg ggcctcggtg
841 cacatgcttt acatgtgttt agtcgaggtt aaaaaacgtc taggcccccc gaaccacggg
901 gacgtggttt tcctttgaaa aacacgatga taatatggcc acaaccatgg tgagcaaggg
961 cgaggagctg ttcaccgggg tggtgcccat cctggtcgag ctggacggcg acgtaaacgg
1021 ccacaagttc agcgtgtccg gcgagggcga gggcgatgcc acctacggca agctgaccct
1081 gaagttcatc tgcaccaccg gcaagctgcc cgtgccctgg cccaccctcg tgaccaccct
1141 gacctacggc gtgcagtgct tcagccgcta ccccgaccac atgaagcagc acgacttctt
1201 caagtccgcc atgcccgaag gctacgtcca ggagcgcacc atcttcttca aggacgacgg
1261 caactacaag acccgcgccg aggtgaagtt cgagggcgac accctggtga accgcatcga
1321 gctgaagggc atcgacttca aggaggacgg caacatcctg gggcacaagc tggagtacaa
1381 ctacaacagc cacaacgtct atatcatggc cgacaagcag aagaacggca tcaaggtgaa
1441 cttcaagatc cgccacaaca tcgaggacgg cagcgtgcag ctcgccgacc actaccagca
1501 gaacaccccc atcggcgacg gccccgtgct gctgcccgac aaccactacc tgagcaccca
1561 gtccgccctg agcaaagacc ccaacgagaa gcgcgatcac atggtcctgc tggagttcgt
1621 gaccgccgcc gggatcactc tcggcatgga cgagctgtac aagtaaggat ccaatcaacc
1681 tctggattac aaaatttgtg aaagattgac tggtattctt aactatgttg ctccttttac
1741 gctatgtgga tacgctgctt taatgccttt gtatcatgct attgcttccc gtatggcttt
1801 cattttctcc tccttgtata aatcctggtt gctgtctctt tatgaggagt tgtggcccgt
1861 tgtcaggcaa cgtggcgtgg tgtgcactgt gtttgctgac gcaaccccca ctggttgggg
1921 cattgccacc acctgtcagc tcctttccgg gactttcgct ttccccctcc ctattgccac
1981 ggcggaactc atcgccgcct gccttgcccg ctgctggaca ggggctcggc tgttgggcac
2041 tgacaattcc gtggtgttgt cggggaaatc atcgtccttt ccttggctgc tcgcctgtgt
2101 tgccacctgg attctgcgcg ggacgtcctt ctgctacaat ccagcggacc ttccttcccg
2161 cggcctgctg ccggctctgc ggcctcttcc gcgtcttcga agtaaaaaaa aaaaaaaaaa
2221 aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa
2281 aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa
2341 aaaaaaaaaa aaaaaaaaaa aaaaaaaaaa ctgccatcag tcggcgtgga ctgtagaaca
2401 ctgccaatgc cggtcccaag cccggataaa agtggagggt acagtccacg ctttttttct
2461 cgagttaagg gcgaattccc gataaggatc ttcctagagc atggctacgt agataagtag
2521 catggcgggt taatcattaa ctacaaggaa cccctagtga tggagttggc cactccctct
2581 ctgcgcgctc gctcgctcac tgaggccggg cgaccaaagg tcgcccgacg cccgggcttt
2641 gcccgggcgg cctcagtgag cgagcgagcg cgcagcctta attaacctaa ttcactggcc
2701 gtcgttttac aacgtcgtga ctgggaaaac cctggcgtta cccaacttaa tcgccttgca
2761 gcacatcccc ctttcgccag ctggcgtaat agcgaagagg cccgcaccga tcgcccttcc
2821 caacagttgc gcagcctgaa tggcgaatgg gacgcgccct gtagcggcgc attaagcgcg
2881 gcgggtgtgg tggttacgcg cagcgtgacc gctacacttg ccagcgccct agcgcccgct
2941 cctttcgctt tcttcccttc ctttctcgcc acgttcgccg gctttccccg tcaagctcta
3001 aatcgggggc tccctttagg gttccgattt agtgctttac ggcacctcga ccccaaaaaa
3061 cttgattagg gtgatggttc acgtagtggg ccatcgccct gatagacggt ttttcgccct
3121 ttgacgttgg agtccacgtt ctttaatagt ggactcttgt tccaaactgg aacaacactc
3181 aaccctatct cggtctattc ttttgattta taagggattt tgccgatttc ggcctattgg
3241 ttaaaaaatg agctgattta acaaaaattt aacgcgaatt ttaacaaaat attaacgctt
3301 acaatttagg tggcactttt cggggaaatg tgcgcggaac ccctatttgt ttatttttct
3361 aaatacattc aaatatgtat ccgctcatga gacaataacc ctgataaatg cttcaataat
3421 attgaaaaag gaagagtatg agtattcaac atttccgtgt cgcccttatt cccttttttg
3481 cggcattttg ccttcctgtt tttgctcacc cagaaacgct ggtgaaagta aaagatgctg
3541 aagatcagtt gggtgcacga gtgggttaca tcgaactgga tctcaacagc ggtaagatcc
3601 ttgagagttt tcgccccgaa gaacgttttc caatgatgag cacttttaaa gttctgctat
3661 gtggcgcggt attatcccgt attgacgccg ggcaagagca actcggtcgc cgcatacact
3721 attctcagaa tgacttggtt gagtactcac cagtcacaga aaagcatctt acggatggca
3781 tgacagtaag agaattatgc agtgctgcca taaccatgag tgataacact gcggccaact
3841 tacttctgac aacgatcgga ggaccgaagg agctaaccgc ttttttgcac aacatggggg
3901 atcatgtaac tcgccttgat cgttgggaac cggagctgaa tgaagccata ccaaacgacg
3961 agcgtgacac cacgatgcct gtagcaatgg caacaacgtt gcgcaaacta ttaactggcg
4021 aactacttac tctagcttcc cggcaacaat taatagactg gatggaggcg gataaagttg
4081 caggaccact tctgcgctcg gcccttccgg ctggctggtt tattgctgat aaatctggag
4141 ccggtgagcg tgggtctcgc ggtatcattg cagcactggg gccagatggt aagccctccc
4201 gtatcgtagt tatctacacg acggggagtc aggcaactat ggatgaacga aatagacaga
4261 tcgctgagat aggtgcctca ctgattaagc attggtaact gtcagaccaa gtttactcat
4321 atatacttta gattgattta aaacttcatt tttaatttaa aaggatctag gtgaagatcc
4381 tttttgataa tctcatgacc aaaatccctt aacgtgagtt ttcgttccac tgagcgtcag
4441 accccgtaga aaagatcaaa ggatcttctt gagatccttt ttttctgcgc gtaatctgct
4501 gcttgcaaac aaaaaaacca ccgctaccag cggtggtttg tttgccggat caagagctac
4561 caactctttt tccgaaggta actggcttca gcagagcgca gataccaaat actgttcttc
4621 tagtgtagcc gtagttaggc caccacttca agaactctgt agcaccgcct acatacctcg
4681 ctctgctaat cctgttacca gtggctgctg ccagtggcga taagtcgtgt cttaccgggt
4741 tggactcaag acgatagtta ccggataagg cgcagcggtc gggctgaacg gggggttcgt
4801 gcacacagcc cagcttggag cgaacgacct acaccgaact gagataccta cagcgtgagc
4861 tatgagaaag cgccacgctt cccgaaggga gaaaggcgga caggtatccg gtaagcggca
4921 gggtcggaac aggagagcgc acgagggagc ttccaggggg aaacgcctgg tatctttata
4981 gtcctgtcgg gtttcgccac ctctgacttg agcgtcgatt tttgtgatgc tcgtcagggg
5041 ggcggagcct atggaaaaac gccagcaacg cggccttttt acggttcctg gccttttgct
5101 ggccttttgc tcacatgttc tttcctgcgt tatcccctga ttctgtggat aaccgtatta
5161 ccgcctttga gtgagctgat accgctcgcc gcagccgaac gaccgagcgc agcgagtcag
5221 tgagcgagga agcggaagag cgcccaatac gcaaaccgcc tctccccgcg cgttggccga
5281 ttcattaatg cagctggcac gacaggtttc ccgactggaa agcgggcagt gagcgcaacg
5341 caattaatgt gagttagctc actcattagg caccccaggc tttacacttt atgcttccgg
5401 ctcgtatgtt gtgtggaatt gtgagcggat aacaatttca cacaggaaac agctatgacc
5461 atgattacgc cagatttaat taagg
//
Circular-ic-EMCV-GFP载体图谱质粒图谱pdf版和Circular-ic-EMCV-GFP载体序列质粒序列等相关资料下载
Circular-ic-EMCV-GFP质粒载体应用举例
- 上一篇:Circular-ic-CVB3-GFP
- 下一篇:没有了

