| 出品公司: | Clontech |
|---|---|
| 载体名称: | pTimer |
| 质粒类型: | 无启动子载体;荧光报告载体 |
| 高拷贝/低拷贝: | 高拷贝 |
| 克隆方法: | 限制性内切酶,多克隆位点 |
| 启动子: | 无启动子 |
| 载体大小: | 4110 bp |
| 5' 测序引物及序列: | DsRed1-N Sequencing Primer (#6482-1) |
| 3' 测序引物及序列: | -- |
| 载体标签: | DsRed1-E5 |
| 载体抗性: | 卡那霉素 |
| 筛选标记: | 新霉素(Neomycin) |
| 克隆菌株: | DH5α |
| 宿主细胞(系): | 真核细胞 |
| 备注: |
pTimer载体不含启动子,将待研究的目的启动子插入MCS区,用于研究启动子或其它顺式转录元件在细胞内的作用; pTimer含有DsRed1-E5荧光报告基因,在表达初始阶段表现为绿色荧光,随着时间的推移,转变为红色荧光; 起到细胞时钟的作用。 |
| 产品目录号: | 632403 |
| 稳定性: | 瞬表达 或 稳表达 |
| 组成型/诱导型: | -- |
| 病毒/非病毒: | 非病毒 |
买家导航

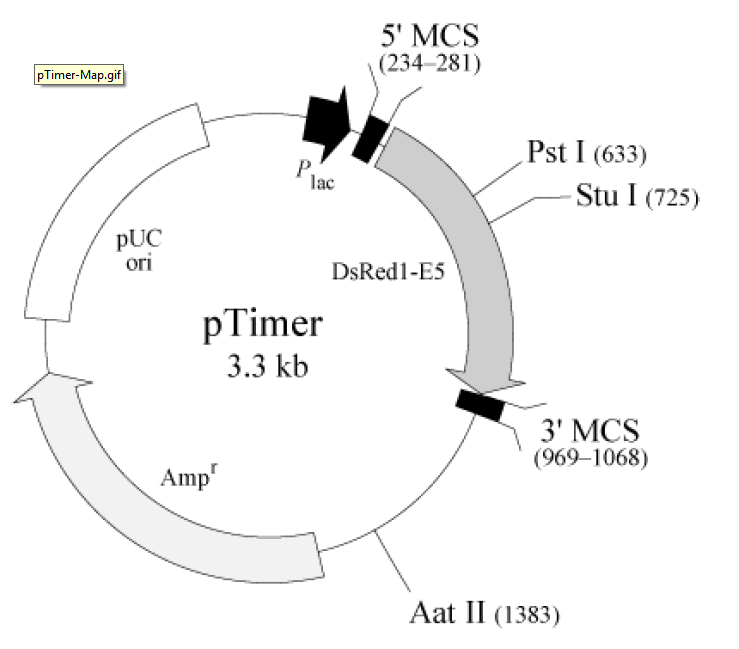
pTimer载体质粒基本信息
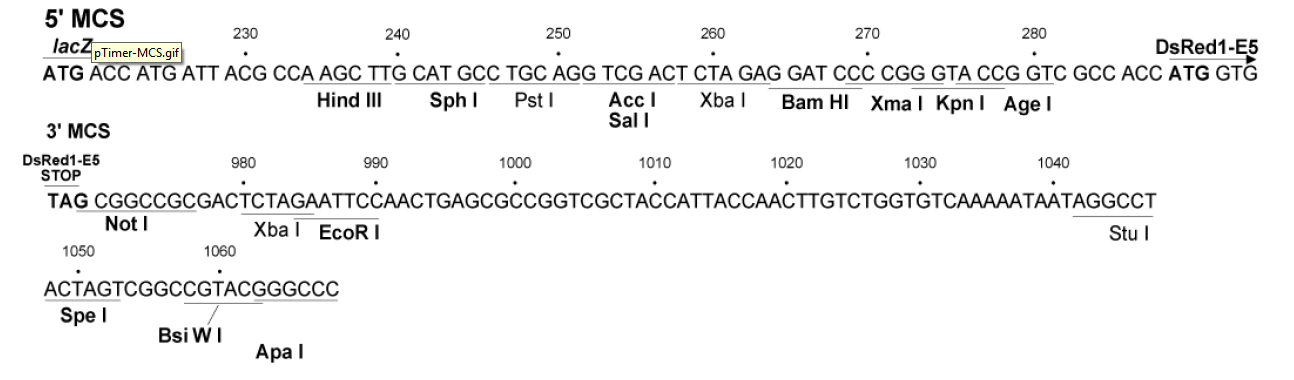
pTimer质粒图谱载体图谱和pTimer载体序列质粒序列多克隆位点信息
pTimer质粒载体简介
pTimer载体描述
pTimer is a prokaryotic expression vector that encodes DsRed1-E5, a fluorescent protein that changes color as it ages. Shortly after translation, DsRed1-E5 emits green light; its red fluorophore emerges later, hours after translation. Because of its predictable color shift, DsRed1-E5 acts as a timer, useful for monitoring changes in gene activity in living cells.
DsRed1-E5 is a mutant of the red fluorescent protein, DsRed1 (1). The cDNA for the wild-type protein, DsRed, was originally isolated by Matz et al., who refer to the protein as drFP583 (2). DsRed1-E5 contains two amino acid substitutions (V105A and S197T), which increase its fluorescence intensity and endow it with a distinct spectral property: As the protein ages, it changes color (3). When first synthesized, DsRed1-E5 is bright green (excitation & emission maxima = 483 nm & 500 nm). As time passes, the green fluorophore undergoes additional changes that cause its fluorescence to shift to longer wavelengths—when fully matured, the protein is bright red (excitation & emission maxima = 558 nm & 583 nm). In mammalian cells transfected with a Tet-inducible DsRed1-E5 expression vector, the green-to-red transition starts about 3 hours after the protein first becomes fluorescent (3). In addition to the amino acid replacements, DsRed1-E5’s coding sequence contains a series of silent base-pair changes, which correspond to human codon-usage preferences, for high expression in mammalian cells (4).
pTimer is primarily intended to serve as a convenient source of the DsRed1-E5 cDNA. The DsRed1-E5 coding sequence is flanked by separate and distinct multiple cloning sites at the 5' and 3' ends so that the gene can easily be excised for use in other expression systems. Alternatively, the DsRed1-E5 coding sequence can be amplified by PCR.
The DsRed1-E5 gene was inserted in frame with the lacZ initiation codon from pUC19 so that DsRed1-E5 is expressed from the lac promoter (Plac) in E. coli host cells. A Kozak consensus sequence is located immediately upstream of DsRed1-E5 to enhance translational efficiency should you wish to express the gene in eukaryotic systems (5). The entire DsRed1-E5 expression cassette in pTimer is supported by a pUC backbone, which contains a high-copy number origin of replication and an ampicillin resistance gene for propagation and selection in E. coli.
When expressed in multicellular organisms, DsRed1-E5’s invariable green-to-red shift can be used as a timer to track the on-off phases of gene expression during embryogenesis and cell differentiation (3).
Recommended Primer Locations: 472–492 and 884–907 (DsRed1-C Sequencing Primer, Cat No. 632388, can be used at 884–907)
Propagation in E. coli Recommended host strain: DH5α Selectable marker: plasmid confers resistance to ampicillin (50 μg/ml) to E. coli hosts. E. coli replication origin: pUC Copy number: high
pTimer质粒序列载体序列
LOCUS pTimer 3314 bp DNA circular SYN 07-MAR-2024
DEFINITION Vector for expressing the Fluorescent Timer in bacteria.
ACCESSION .
VERSION .
KEYWORDS pTimer
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 3314)
AUTHORS Clontech (TaKaRa)
TITLE Direct Submission
JOURNAL Exported May 12, 2025 from SnapGene 6.0.2
https://www.snapgene.com
COMMENT Translation from the lacZ start codon produces a fusion protein in
E. coli.
FEATURES Location/Qualifiers
source 1..3314
/lab_host="Escherichia coli"
/mol_type="other DNA"
/organism="synthetic DNA construct"
promoter 143..173
/label=lac promoter
/note="promoter for the E. coli lac operon"
protein_bind 181..197
/label=lac operator
/bound_moiety="lac repressor encoded by lacI"
/note="The lac repressor binds to the lac operator to
inhibit transcription in E. coli. This inhibition can be
relieved by adding lactose or
isopropyl-beta-D-thiogalactopyranoside (IPTG)."
CDS 217..219
/codon_start=1
/product="lacZ start codon"
/label=ATG
/translation="M"
CDS 289..969
/codon_start=1
/product="green-to-red fluorescent timer derivative of
DsRed"
/label=Timer
/note="mammalian codon-optimized"
/translation="MVRSSKNVIKEFMRFKVRMEGTVNGHEFEIEGEGEGRPYEGHNTV
KLKVTKGGPLPFAWDILSPQFQYGSKVYVKHPADIPDYKKLSFPEGFKWERVMNFEDGG
VATVTQDSSLQDGCFIYKVKFIGVNFPSDGPVMQKKTMGWEASTERLYPRDGVLKGEIH
KALKLKDGGHYLVEFKSIYMAKKPVQLPGYYYVDTKLDITSHNEDYTIVEQYERTEGRH
HLFL"
promoter 1409..1513
/gene="bla"
/label=AmpR promoter
CDS 1514..2374
/codon_start=1
/gene="bla"
/product="beta-lactamase"
/label=AmpR
/note="confers resistance to ampicillin, carbenicillin, and
related antibiotics"
/translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGYI
ELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRIDAGQEQLGRRIHYSQNDLVEYS
PVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRLDRW
EPELNEAIPNDERDTTMPVAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPLLRSA
LPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIAEIGAS
LIKHW"
rep_origin 2545..3133
/direction=RIGHT
/label=ori
/note="high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
ORIGIN
1 agcgcccaat acgcaaaccg cctctccccg cgcgttggcc gattcattaa tgcagctggc
61 acgacaggtt tcccgactgg aaagcgggca gtgagcgcaa cgcaattaat gtgagttagc
121 tcactcatta ggcaccccag gctttacact ttatgcttcc ggctcgtatg ttgtgtggaa
181 ttgtgagcgg ataacaattt cacacaggaa acagctatga ccatgattac gccaagcttg
241 catgcctgca ggtcgactct agaggatccc cgggtaccgg tcgccaccat ggtgcgctcc
301 tccaagaacg tcatcaagga gttcatgcgc ttcaaggtgc gcatggaggg caccgtgaac
361 ggccacgagt tcgagatcga gggcgagggc gagggccgcc cctacgaggg ccacaacacc
421 gtgaagctga aggtgaccaa gggcggcccc ctgcccttcg cctgggacat cctgtccccc
481 cagttccagt acggctccaa ggtgtacgtg aagcaccccg ccgacatccc cgactacaag
541 aagctgtcct tccccgaggg cttcaagtgg gagcgcgtga tgaacttcga ggacggcggc
601 gtggcgaccg tgacccagga ctcctccctg caggacggct gcttcatcta caaggtgaag
661 ttcatcggcg tgaacttccc ctccgacggc cccgtgatgc agaagaagac catgggctgg
721 gaggcctcca ccgagcgcct gtacccccgc gacggcgtgc tgaagggcga gatccacaag
781 gccctgaagc tgaaggacgg cggccactac ctggtggagt tcaagtccat ctacatggcc
841 aagaagcccg tgcagctgcc cggctactac tacgtggaca ccaagctgga catcacctcc
901 cacaacgagg actacaccat cgtggagcag tacgagcgca ccgagggccg ccaccacctg
961 ttcctgtagc ggccgcgact ctagaattcc aactgagcgc cggtcgctac cattaccaac
1021 ttgtctggtg tcaaaaataa taggcctact agtcggccgt acgggccctt tcgtctcgcg
1081 cgtttcggtg atgacggtga aaacctctga cacatgcagc tcccggagac ggtcacagct
1141 tgtctgtaag cggatgccgg gagcagacaa gcccgtcagg gcgcgtcagc gggtgttggc
1201 gggtgtcggg gctggcttaa ctatgcggca tcagagcaga ttgtactgag agtgcaccat
1261 atgcggtgtg aaataccgca cagatgcgta aggagaaaat accgcatcag gcggccttaa
1321 gggcctcgtg atacgcctat ttttataggt taatgtcatg ataataatgg tttcttagac
1381 gtcaggtggc acttttcggg gaaatgtgcg cggaacccct atttgtttat ttttctaaat
1441 acattcaaat atgtatccgc tcatgagaca ataaccctga taaatgcttc aataatattg
1501 aaaaaggaag agtatgagta ttcaacattt ccgtgtcgcc cttattccct tttttgcggc
1561 attttgcctt cctgtttttg ctcacccaga aacgctggtg aaagtaaaag atgctgaaga
1621 tcagttgggt gcacgagtgg gttacatcga actggatctc aacagcggta agatccttga
1681 gagttttcgc cccgaagaac gttttccaat gatgagcact tttaaagttc tgctatgtgg
1741 cgcggtatta tcccgtattg acgccgggca agagcaactc ggtcgccgca tacactattc
1801 tcagaatgac ttggttgagt actcaccagt cacagaaaag catcttacgg atggcatgac
1861 agtaagagaa ttatgcagtg ctgccataac catgagtgat aacactgcgg ccaacttact
1921 tctgacaacg atcggaggac cgaaggagct aaccgctttt ttgcacaaca tgggggatca
1981 tgtaactcgc cttgatcgtt gggaaccgga gctgaatgaa gccataccaa acgacgagcg
2041 tgacaccacg atgcctgtag caatggcaac aacgttgcgc aaactattaa ctggcgaact
2101 acttactcta gcttcccggc aacaattaat agactggatg gaggcggata aagttgcagg
2161 accacttctg cgctcggccc ttccggctgg ctggtttatt gctgataaat ctggagccgg
2221 tgagcgtggg tctcgcggta tcattgcagc actggggcca gatggtaagc cctcccgtat
2281 cgtagttatc tacacgacgg ggagtcaggc aactatggat gaacgaaata gacagatcgc
2341 tgagataggt gcctcactga ttaagcattg gtaactgtca gaccaagttt actcatatat
2401 actttagatt gatttaaaac ttcattttta atttaaaagg atctaggtga agatcctttt
2461 tgataatctc atgaccaaaa tcccttaacg tgagttttcg ttccactgag cgtcagaccc
2521 cgtagaaaag atcaaaggat cttcttgaga tccttttttt ctgcgcgtaa tctgctgctt
2581 gcaaacaaaa aaaccaccgc taccagcggt ggtttgtttg ccggatcaag agctaccaac
2641 tctttttccg aaggtaactg gcttcagcag agcgcagata ccaaatactg ttcttctagt
2701 gtagccgtag ttaggccacc acttcaagaa ctctgtagca ccgcctacat acctcgctct
2761 gctaatcctg ttaccagtgg ctgctgccag tggcgataag tcgtgtctta ccgggttgga
2821 ctcaagacga tagttaccgg ataaggcgca gcggtcgggc tgaacggggg gttcgtgcac
2881 acagcccagc ttggagcgaa cgacctacac cgaactgaga tacctacagc gtgagctatg
2941 agaaagcgcc acgcttcccg aagggagaaa ggcggacagg tatccggtaa gcggcagggt
3001 cggaacagga gagcgcacga gggagcttcc agggggaaac gcctggtatc tttatagtcc
3061 tgtcgggttt cgccacctct gacttgagcg tcgatttttg tgatgctcgt caggggggcg
3121 gagcctatgg aaaaacgcca gcaacgcggc ctttttacgg ttcctggcct tttgctggcc
3181 ttttgctcac atgttctttc ctgcgttatc ccctgattct gtggataacc gtattaccgc
3241 ctttgagtga gctgataccg ctcgccgcag ccgaacgacc gagcgcagcg agtcagtgag
3301 cgaggaagcg gaag
//
pTimer载体图谱质粒图谱pdf版和pTimer载体序列质粒序列等相关资料下载
pTimer质粒载体应用举例
- 上一篇:pHcRed1-DR
- 下一篇:pHcRed1-1